The EM algorithm
For many models in machine learning and statistics, computing the ML or MAP parameter estimate is easy provided we observe all the values of all the relevant random variables, i.e., if we have complete data. However, if we have missing data and/or latent variables, then computing the ML/MAP estimate becomes hard. One approach is to use a generic gradient-based optimizer to find a local minimum of the negative log likelihood or NLL, given by
However, we often have to enforce constraints, such as the fact that covariance matrices must be positive definite, mixing weights must sum to one, etc., which can be tricky. In such cases, it is often much simpler (but not always faster) to use an algorithm called expectation maximization, or EM for short. This is a simple iterative algorithm, often with closed-form updates at each step. Furthermore, the algorithm automatically enforce the required constraints. EM exploits the fact that if the data were fully observed, then the ML/ MAP estimate would be easy to compute. In particular, EM is an iterative algorithm which alternates between inferring the missing values given the parameters (E step), and then optimizing the parameters given the “filled in” data (M step). We give the details below, followed by several examples.
Let
Unfortunately this is hard to optimize, since the log cannot be pushed inside the sum.
EM gets around this problem as follows. Define the complete data log likelihood to be
Let the marginal likelihood as
The complete data log likelihod cannot be computed. So lets define the expected complete data log likelihood as follows:
To perform MAP to avoid overfitting:
We could easily maximize
E-step:
Taking maxture of Gausians as an example, the expected complete data log likelihood is given by
where
M step: in the M step, we optimize Q wrt
Which is the averaged responsibility for cluster
This is just a weighted version of the standard problem of computing the MLEs of an MVN and the new parameter estimate is given by
import numpy as np
def gaussian_point_generator(mu:list,sigma:list,n:int):
return list(zip(np.random.normal(mu[0], sigma[0], n), np.random.normal(mu[1], sigma[1], n)))
class GaussianParameter:
def __init__(self, mu:list,sigma:list, cov=0):
self.mu = mu
self.sigma = sigma
self.cov=cov
def __repr__(self):
return "".format(self.mu, self.sigma)
def plot(self, ax):
m = np.array([[self.mu[0]],[self.mu[1]]])
cov = np.array(self.sigma)
cov_inv = np.linalg.inv(cov) # inverse of covariance matrix
cov_det = np.linalg.det(cov) # determinant of covariance matrix
x = np.linspace(-20,20)
y = np.linspace(-20,20)
X,Y = np.meshgrid(x,y)
coe = 1.0 / ((2 * np.pi)**2 * cov_det)**0.5
Z = coe * np.e ** (-0.5 * (cov_inv[0,0]*(X-m[0])**2 + (cov_inv[0,1] + cov_inv[1,0])*(X-m[0])*(Y-m[1]) + cov_inv[1,1]*(Y-m[1])**2))
ax.contour(X,Y,Z)
def p(self, point):
return np.array((1 / 2 * np.pi * np.power(np.linalg.det(np.matrix(self.sigma)), 0.5) ) * np.exp( -1/2 * (np.matrix(np.array(self.mu)) - np.matrix(np.array(point))) * np.matrix(self.sigma) * (np.matrix(np.array(self.mu)) - np.matrix(np.array(point))).T))[0][0]
init_gaussian_parameter = [GaussianParameter([0,10], [[1,0],[0,1]]), GaussianParameter([10,0], [[1,0],[0,1]])]
points = gaussian_point_generator([1,1], [1,1],50) + gaussian_point_generator([9,9], [2,2],50)
def iterate(axis):
r = [[], []]
for i in points:
for j in range(2):
r[j].append(init_gaussian_parameter[j].p(i))
r[0], r[1] = np.array(r[0]) / (np.array(r[0]) + np.array(r[1])), np.array(r[1]) / (np.array(r[0]) + np.array(r[1]))
r[0] = np.array(list(map(lambda x:0 if np.isnan(x) else x, r[0])))
r[1] = np.array(list(map(lambda x:0 if np.isnan(x) else x, r[1])))
new_mu = [np.array([0,0]), np.array([0,0])]
for j in range(2):
for ind,i in enumerate(points):
new_mu[j] = new_mu[j] + r[j][ind] * np.array(i)
new_mu[j] = new_mu[j] / np.sum(r[j])
new_sigma = [np.array([[0,0], [0,0]]), np.array([[0,0], [0,0]])]
for j in range(2):
for ind,i in enumerate(points):
new_sigma[j] = new_sigma[j] + r[j][ind] * np.matrix(i).T * np.matrix(i)
new_sigma[j] = new_sigma[j] / np.sum(r[j])
new_sigma[j] = new_sigma[j] - np.matrix(new_mu[j]).T * np.matrix(new_mu[j])
init_gaussian_parameter[j].mu = np.array(new_mu[j])
init_gaussian_parameter[j].sigma = np.array(new_sigma[j])
init_gaussian_parameter[0].plot(axis)
init_gaussian_parameter[1].plot(axis)
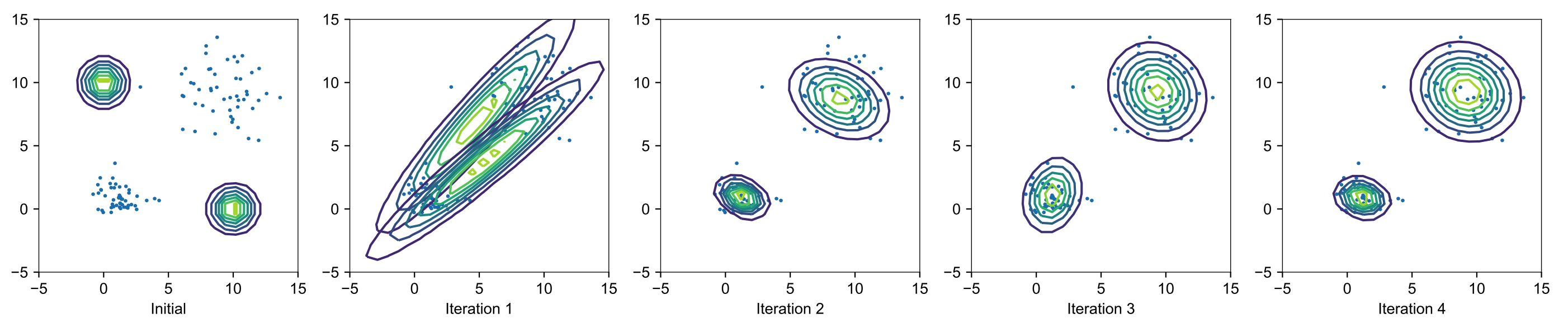
The result gives